MARS Regression in R
06.23.2021
Intro
Multivariate Adaptive Regression Splines or MARS is a regression model that extends linear models to nonlinear. It essentially creates many piecewise functions to model your data. In this article, we will learn how to use MARS Regression in R.
Data
For this tutorial, we will use the Boston data set which includes
housing data with features of the houses and their prices. We would like
to predict the medv column or the medium value.
library(MASS)
data(Boston)
str(Boston)## 'data.frame': 506 obs. of 14 variables:
## $ crim : num 0.00632 0.02731 0.02729 0.03237 0.06905 ...
## $ zn : num 18 0 0 0 0 0 12.5 12.5 12.5 12.5 ...
## $ indus : num 2.31 7.07 7.07 2.18 2.18 2.18 7.87 7.87 7.87 7.87 ...
## $ chas : int 0 0 0 0 0 0 0 0 0 0 ...
## $ nox : num 0.538 0.469 0.469 0.458 0.458 0.458 0.524 0.524 0.524 0.524 ...
## $ rm : num 6.58 6.42 7.18 7 7.15 ...
## $ age : num 65.2 78.9 61.1 45.8 54.2 58.7 66.6 96.1 100 85.9 ...
## $ dis : num 4.09 4.97 4.97 6.06 6.06 ...
## $ rad : int 1 2 2 3 3 3 5 5 5 5 ...
## $ tax : num 296 242 242 222 222 222 311 311 311 311 ...
## $ ptratio: num 15.3 17.8 17.8 18.7 18.7 18.7 15.2 15.2 15.2 15.2 ...
## $ black : num 397 397 393 395 397 ...
## $ lstat : num 4.98 9.14 4.03 2.94 5.33 ...
## $ medv : num 24 21.6 34.7 33.4 36.2 28.7 22.9 27.1 16.5 18.9 ...Basic MARS Regression Model in R
library(earth)## Warning: package 'earth' was built under R version 4.0.5
## Loading required package: Formula
## Loading required package: plotmo
## Warning: package 'plotmo' was built under R version 4.0.5
## Loading required package: plotrix
## Loading required package: TeachingDemos
## Warning: package 'TeachingDemos' was built under R version 4.0.5model = earth(medv ~ ., data = Boston)
model## Selected 20 of 24 terms, and 10 of 13 predictors
## Termination condition: Reached nk 27
## Importance: rm, lstat, ptratio, dis, nox, crim, tax, rad, black, indus, ...
## Number of terms at each degree of interaction: 1 19 (additive model)
## GCV 12.44743 RSS 5364.917 GRSq 0.853135 RSq 0.8744059Modeling Ridge Regression in R with Caret
We will now see how to model a ridge regression using the Caret
package. We will use this library as it provides us with many features
for real life modeling.
To do this, we use the train method. We pass the same parameters as
above, but in addition we pass the method = 'ridge' model to tell
Caret to use a lasso model.
library(caret)## Loading required package: lattice
## Loading required package: ggplot2set.seed(1)
model <- train(
medv ~ .,
data = Boston,
method = 'earth'
)
model## Multivariate Adaptive Regression Spline
##
## 506 samples
## 13 predictor
##
## No pre-processing
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 506, 506, 506, 506, 506, 506, ...
## Resampling results across tuning parameters:
##
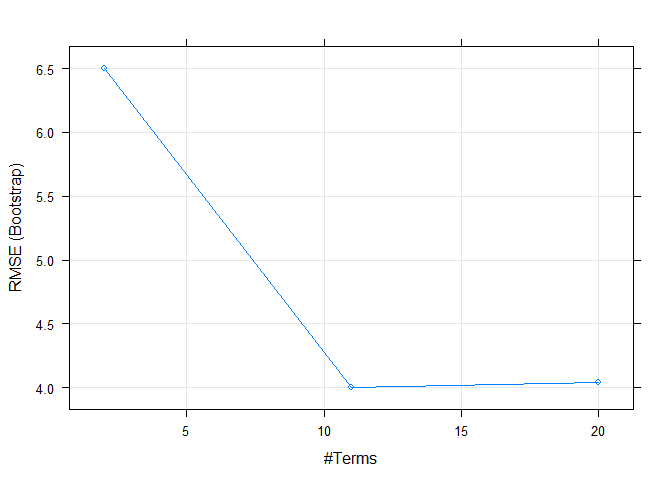
## nprune RMSE Rsquared MAE
## 2 6.497277 0.5070059 4.659254
## 11 4.005270 0.8098340 2.775501
## 20 4.044197 0.8076821 2.767983
##
## Tuning parameter 'degree' was held constant at a value of 1
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nprune = 11 and degree = 1.Here we can see that caret automatically trained over multiple hyper parameters. We can easily plot those to visualize.
plot(model)Preprocessing with Caret
One feature that we use from Caret is preprocessing. Often in real life
data science we want to run some pre processing before modeling. We will
center and scale our data by passing the following to the train method:
preProcess = c("center", "scale").
set.seed(1)
model2 <- train(
medv ~ .,
data = Boston,
method = 'earth',
preProcess = c("center", "scale")
)
model2## Multivariate Adaptive Regression Spline
##
## 506 samples
## 13 predictor
##
## Pre-processing: centered (13), scaled (13)
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 506, 506, 506, 506, 506, 506, ...
## Resampling results across tuning parameters:
##
## nprune RMSE Rsquared MAE
## 2 6.497277 0.5070059 4.659254
## 11 4.005270 0.8098340 2.775501
## 20 4.044129 0.8076882 2.767972
##
## Tuning parameter 'degree' was held constant at a value of 1
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nprune = 11 and degree = 1.Splitting the Data Set
Often when we are modeling, we want to split our data into a train and
test set. This way, we can check for overfitting. We can use the
createDataPartition method to do this. In this example, we use the
target medv to split into an 80/20 split, p = .80.
This function will return indexes that contains 80% of the data that we should use for training. We then use the indexes to get our training data from the data set.
set.seed(1)
inTraining <- createDataPartition(Boston$medv, p = .80, list = FALSE)
training <- Boston[inTraining,]
testing <- Boston[-inTraining,]We can then fit our model again using only the training data.
set.seed(1)
model3 <- train(
medv ~ .,
data = training,
method = 'earth',
preProcess = c("center", "scale")
)
model3## Multivariate Adaptive Regression Spline
##
## 407 samples
## 13 predictor
##
## Pre-processing: centered (13), scaled (13)
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 407, 407, 407, 407, 407, 407, ...
## Resampling results across tuning parameters:
##
## nprune RMSE Rsquared MAE
## 2 6.512199 0.5019664 4.669863
## 11 4.351181 0.7725451 2.955287
## 20 4.300871 0.7803163 2.886865
##
## Tuning parameter 'degree' was held constant at a value of 1
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nprune = 20 and degree = 1.Now, we want to check our data on the test set. We can use the subset
method to get the features and test target. We then use the predict
method passing in our model from above and the test features.
Finally, we calculate the RMSE and r2 to compare to the model above.
test.features = subset(testing, select=-c(medv))
test.target = subset(testing, select=medv)[,1]
predictions = predict(model3, newdata = test.features)
# RMSE
sqrt(mean((test.target - predictions)^2))## [1] 3.570804# R2
cor(test.target, predictions) ^ 2## y
## [1,] 0.855721Cross Validation
In practice, we don’t normal build our data in on training set. It is
common to use a data partitioning strategy like k-fold cross-validation
that resamples and splits our data many times. We then train the model
on these samples and pick the best model. Caret makes this easy with the
trainControl method.
We will use 10-fold cross-validation in this tutorial. To do this we
need to pass three parameters method = "cv", number = 10 (for
10-fold). We store this result in a variable.
set.seed(1)
ctrl <- trainControl(
method = "cv",
number = 10,
)Now, we can retrain our model and pass the trainControl response to
the trControl parameter. Notice the our call has added
trControl = set.seed.
# set.seed(1)
model4 <- train(
medv ~ .,
data = training,
method = 'earth',
# preProcess = c("center", "scale"),
trControl = trainControl(method="cv"),
)
model4## Multivariate Adaptive Regression Spline
##
## 407 samples
## 13 predictor
##
## No pre-processing
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 367, 366, 367, 366, 365, 367, ...
## Resampling results across tuning parameters:
##
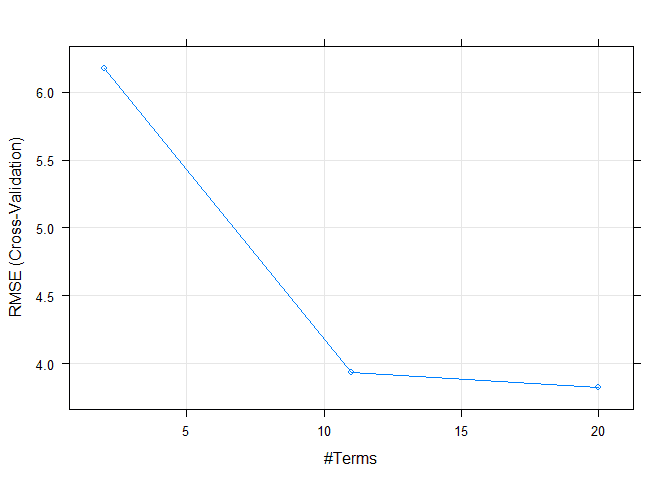
## nprune RMSE Rsquared MAE
## 2 6.172609 0.5477370 4.504196
## 11 3.933227 0.8191702 2.794837
## 20 3.827388 0.8237172 2.780469
##
## Tuning parameter 'degree' was held constant at a value of 1
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nprune = 20 and degree = 1.plot(model4)This results seemed to have improved our accuracy for our training data. Let’s check this on the test data to see the results.
test.features = subset(testing, select=-c(medv))
test.target = subset(testing, select=medv)[,1]
predictions = predict(model4, newdata = test.features)
# RMSE
sqrt(mean((test.target - predictions)^2))## [1] 3.570804# R2
cor(test.target, predictions) ^ 2## y
## [1,] 0.855721Tuning Hyper Parameters
To tune a ridge model, we can give the model different values of
lambda. Caret will retrain the model using different lambdas and
select the best version.
set.seed(1)
tuneGrid <- expand.grid(
degree = 1,
nprune = c(2, 11, 10)
)
model5 <- train(
medv ~ .,
data = training,
method = 'earth',
preProcess = c("center", "scale"),
trControl = ctrl,
tuneGrid = tuneGrid
)
model5## Multivariate Adaptive Regression Spline
##
## 407 samples
## 13 predictor
##
## Pre-processing: centered (13), scaled (13)
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 367, 366, 367, 366, 365, 367, ...
## Resampling results across tuning parameters:
##
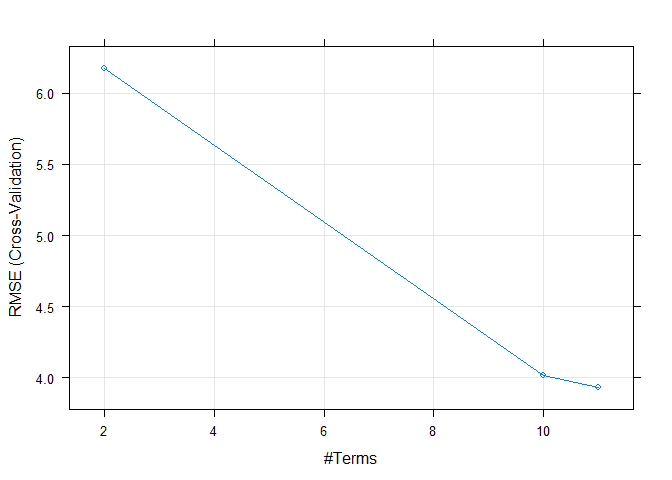
## nprune RMSE Rsquared MAE
## 2 6.172609 0.5477370 4.504196
## 10 4.014690 0.8120098 2.837775
## 11 3.933227 0.8191702 2.794837
##
## Tuning parameter 'degree' was held constant at a value of 1
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were nprune = 11 and degree = 1.Finally, we can again plot the model to see how it performs over different tuning parameters.
plot(model5)